This is not the most exciting blog title I’ve ever come up with but it’s important to teach students how to keep records, especially when collecting lab results that will be used to write a report. In my previous entry, I discussed our involvement in One Brave Idea, which could provide our students with an opportunity to present lab results in an ongoing pAcne study to experts in the field of cardiovascular and microbiome health. Many students were excited by this and wanted to put their best foot forward to impress the scientists but there was something crucial that needed to be taught regarding record-keeping.
When students hand in work, I tend to notice that a fair number of them don’t include pictures they took during the experiment. I know they took them but I realized that they weren’t sure how to keep them readily accessible a week or a month after the experiment. This initially surprised me. After all, how many pictures do we have on our phones at the ready to be shown to others? On the other hand, how many times have you tried to access a specific one to show someone and found yourself aimlessly swiping across your photo roll, only to give up and sigh, “it’s in there somewhere.” If you’re like me, quite a few, so it’s reasonable to assume students would struggle with this problem too. So onwards began our thrilling unit on record keeping!
I started our discussion like an infomercial by saying, “Has this every happened to you: You take a picture of a lab result, look at it proudly and think, ‘this is a good picture. I’m going to use it in the lab report.’ But then during the writing process, the picture is gone, replaced by countless others leaving you high and dry?” Virtually every student nodded their hands and a couple started laughing as they did so. Even kids who had phones on them still lost pictures because they have never had to catalog them.
The question was posited: If we want to hold on pics, what should we do? The students immediately had plenty of ideas which included: emailing them to yourself with a predictable subject (ResultsDateInitials), uploading them to a labelled folder (DateResults) in Drive, Creating a Document called “LabResultsUnitNameDate”, and various iterations of that. After they all had a plan, they went to work, collected their results, and saved them. Some of them made sure to take pictures that showed the labels on the petri dishes to make it even easier for them. The entire module with talk and collection took about 20 minutes but now the students have a means to collect and keep information. I did a brief review of what we learned when we collected a second round of pictures but that was much quicker and it will eventually become a part of our routines.
I wrote earlier this year about executive functioning and the importance of thinking ahead and going through the process of planning backwards from goal to execution. This recap of GLP for record keeping is a good example of scaffolding that most students could use practice in, whether they have stellar EF skills or not. The students will all have to write about this experiment. They will all have to refer to pictures. They all now have a place where they can go to find that quickly saving them time and anguish. A nice bonus is that they shared their folders with me so I can access their photos if I need them (like for this blog post).
Next time, I will highlight how we cultured our pAcne in anaerobic conditions and how the entire unit will lead us into a discussion of microevolution and natural selection.
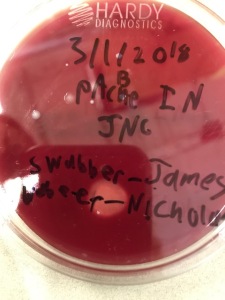
Clearly labeled and visible in pics